About
The IsoPops package is a product of a research project performed by Tom Ray and Kelly Cochran in the Kay Lab at Duke University. The aim of the project is to characterize the isoform diversity of cell-surface receptors expressed in the retina, using PacBio long-read transcriptome sequencing.
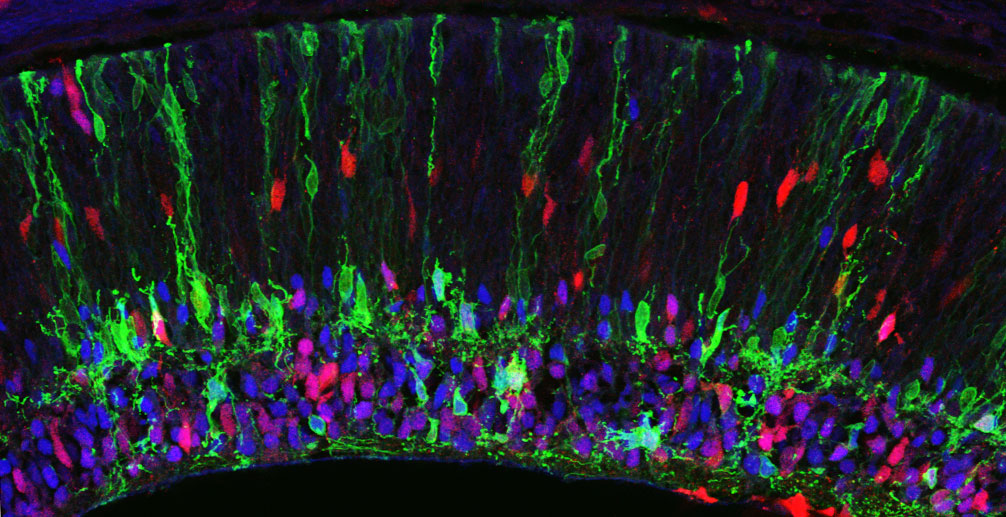
Diverse subtypes of neurons organize into complex spatial mosaics within the retina during development. Exactly how a developing neuron knows to differentiate or localize to its correct subtype or position is not well-understood. Cell-surface receptors may function as a means of communication between neighboring differentiating cells.
Utilizing novel experimental techniques for capturing increased read depth at our target genes, we were able to collect transcripts which provided evidence for thousands of potential unannotated isoforms. From there, we set out to develop a bioinformatics framework to validate, summarize, and visualize this previously unconfirmed transcript diversity.
IsoPops takes in the files produced by PacBio's IsoSeq protocol for transcriptome processing. First, it compiles the information from the IsoSeq protocol into more compact and interpretable tables within a Database object. Database objects can be provided gene identifying information, which allows for the creation of an additional table containing gene-level summary information about the transcriptome. Next, IsoPops optionally filters out transcripts by user-defined length and/or read count cutoffs and removes transcripts which are likely to be sequencing artifacts. From there, any number of analyses can be performed on the data, including vectorization, dimension reduction (PCA), and hierarchical clustering so that isoform diversity can be easily visualized. IsoPops includes extensive graphing functionality through the popular R package ggplot2, letting you generate the plots you want in a consistent, manuscript-quality style. The entire workflow can be run in only a few lines of code, but is designed to allow for thorough customization of analyses and plots if desired.
The manuscript for the project is currently in preparation and expected to be submitted as a preprint to BioArxiv soon. For more information, feel free to contact Tom (ray@neuro.duke.edu), Kelly (kelly.cochran36@gmail.com), or Jeremy Kay (kay@neuro.duke.edu).